Many enzymes catalyze the decarboxylation of organic acids. The mechanisms utilized by biological decarboxylases often involve the formation of covalently bound intermediates that provide a means of stabilizing the anion formed upon loss of carbon dioxide through resonance (such mechanisms often involve cofactors such as thiamin diphosphate and pyridoxal phosphate).
Of particular interest, however, are cases of direct decarboxylation in which no covalent intermediates are formed. Two such examples are described below.
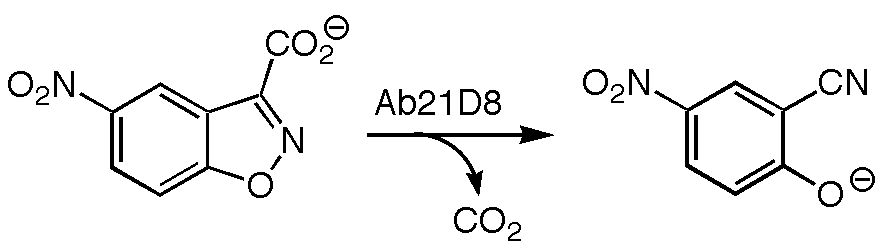
Antibody Catalyzed Decarboxylation of Carboxybenzisoxazoles
The decarboxylation of 3-carboxybenzisoxazoles has been of interest to chemists for decades due to the tremendous acceleration of this reaction in nonpolar and polar aprotic environments versus reaction in aqueous solution. Recently, the structure of an antibody (21D8) that catalyzes the decarboxylation of carboxybenzisoxazoles has been obtained by the research groups of Donald Hilvert at the Laboratory of Organic Chemistry at the Swiss Federal Institute of Technology (ETH) and Ian Wilson in the Department of Molecular Biology and Skaggs Institute for Chemical Biology at the Scripps Research Institute. Based on this crystal structure, and in collaboration with Hilvert and Wilson, a combination of quantum mechanical, automated docking, and molecular dynamics calculations were used to explore the origins of catalysis by antibody 21D8. It appears that this antibody stabilizes the transition state for decarboxylation primarily through specific preorganized noncovalent interactions rather than through more homogeneous medium effects.
Hotta, K.; Lange, H.; Tantillo, D. J.; Houk, K. N.; Hilvert, D.; Wilson, I. A. J. Mol. Biol. 2000, 302, 1213-1225: "Catalysis of Decarboxylation by a Preorganized Heterogenous Microenvironment: Crystal Structures of Abzyme 21D8"
Ujaque, G.; Tantillo, D. J.; Hu, Y.; Houk, K. N.; Hotta, K.; Hilvert, D. J. Comp. Chem. 2002, 24, 98-110: "Catalysis on the Coastline: Theozyme, Molecular Dynamics, and Free Energy Perturbation Analysis of Antibody 21D8 Catalysis of the Decarboxylation of 5-Nitro-3-Carboxybenzisoxazole," part of a special issue honoring Dr. Peter A. Kollman.
Rodrigues, A. V.; Tantillo, D. J.; Mukhopadhyay, A.; Keasling, J. D.; Beller, H. R. ChemBioChem 2020, 21, 663-671: "Insights into the Mechanism of Phenylacetate Decarboxylase (PhdB), a Novel Glycyl Radical Enzyme"
Dean J. Tantillo and K. N. Houk: "Analogies Between Antibody Hydrolases and Decarboxylases" Poster presented at the 5th Annual Maria Goeppert-Mayer Interdisciplinary Symposium, San Diego, CA, March 4, 2000.
Dean J. Tantillo, Kinya Hotta, Donald Hilvert, and K. N. Houk: "Origins of Catalysis and Cross-Reactivity for an Antibody Decarboxylase" Poster presented at the 219th ACS National Meeting, San Francisco, CA, March 26-30, 2000; paper ORGN 422.
Dean J. Tantillo: "Theoretical Bioorganic Chemistry of Antibody Catalysis: Understanding an Antibody Decarboxylase" Lecture presented at the UCLA Department of Chemistry and Biochemistry Organic Chemistry Graduate Symposium, Honoring Christopher S. Foote on His 65th Birthday, Los Angeles, CA, June 3, 2000.
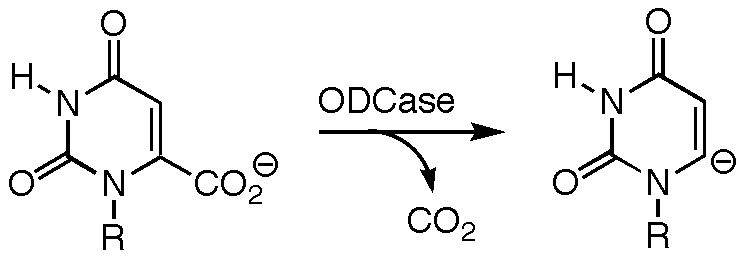
The Mechanism of Orotidine-5'-monophosphate Decarboxylase
The mechanism by which orotodine 5'-monophosphate (OMP) decarboxylase (ODCase) produces uridine 5'-monophosphate (UMP) has been the subject of much heated debate. ODCase is among the most proficient enzymes known, yet not only have no cofactors or covalent intermediates been implicated in the catalytic mechanism, but the negative charge formed upon loss of carbon dioxide from OMP cannot be stabilized through resonance (the breaking C---C bond is orthogonal to the pi-system to which it is attached). Several groups have proposed that catalysis arises primarily from destabilizing interactions between the carboxylate group in OMP and an aspartate residue in the binding site, these unfavorable interactions being relieved as decarboxylation proceeds. Our calculations show, however, that two carboxylates in such close proximity (even in environments with high dielectric constants) have a huge affinity for a proton or a bridging ammonium group. This suggests that the enormous decrease in activation barrier by ODCase does not arise from ground state destabilization, since the substrate and enzyme carboxylates have various avenues to avoid that destabilization. Several other mechanisms have been proposed (see, for example, the work of Jeehiun Lee at Rutgers University), but for now at least, the catalytic conundrum remains.
Houk, K. N.; Lee, J. K.; Tantillo, D. J.; Bahmanyar, S.; Hietbrink, B. N. ChemBioChem 2001, 2, 113-118: "Crystal Structures of Orotidine Monophosphate Decarboxylase: Does the Structure Reveal the Mechanism of the Most Proficient Enzyme?"
Lee, J. K.; Tantillo, D. J. Adv. Phys. Org. Chem. 2003, 38, 183-218 : "Computational Insights into the Mechanism of Orotidine Monophosphate Decarboxylase," part of a volume honoring Prof. Ken Houk on the occasion of his 60th birthday.
Houk, K. N.; Tantillo, D. J.; Stanton, C.; Hu, Y. Top. Curr. Chem. 2004, 238, 1-22: "What has Theory and Crystallography Revealed About the Mechanism of Catalysis by Orotidine Monophosphate Decarboxylase?"
back to list of research projects